We use cookies to enhance your experience. By continuing to browse this site you agree to our use of cookies. More info.
By clicking "Allow All" you agree to the storing of cookies on your device to enhance site navigation, analyse site usage and support us in providing free open access scientific content. More info. Small Rna Extraction Kit
Recent advancements in next-generation sequencing have led to substantial increases in throughput, coupled with reduced expenses.
Due to these improvements, the process of library preparation has become even more of a financial and time constraint to high throughput sequencing core facilities. Nonetheless, with the help of automation in the form of liquid-handling robots, some of these bottlenecks can be alleviated.
This article outlines the utilization of one such system, the mosquito® HV system, to efficiently execute DNA library preparation for Illumina sequencing, reducing the volume to one-tenth of the original manual protocol.
mosquito HV genomics, a fully open liquid handler, offers several advantages:
The mosquito system, developed by SPT Labtech, employs positive displacement for pipetting, utilizing disposable tips containing a small stainless steel rod (see Figure 1).
This pipetting mechanism allows for precise handling of nanoliter volumes, eliminating the need for specialized liquid classifications in the instrument control software.
Figure 1. mosquito positive displacement tip technology and mosquito HV genomics. Image Credit: SPT Labtech
The selection of the NEBNext Ultra II FS DNA method was based on its enzymatic fragmentation capability, eliminating the need for physical DNA fragmentation at the onset of library preparation.
Using ZymoBIOMICS Microbial Community DNA Standard as genomic DNA, the performance of the mosquito HV system was compared with manually prepared libraries at standard volumes, using varying DNA input quantities.
To evaluate outcomes with less standardized input material, reduced-volume NEBNext Ultra II FS DNA libraries were generated from individual bacterial isolates and compared with manually prepared Illumina TruSeq Nano libraries derived from the same input material.
Additionally, a comparison was made between the NEBNext Ultra II FS DNA automated libraries and TruSeq Nano and TruSeq PCR Free libraries produced through manual methods or traditional automation.
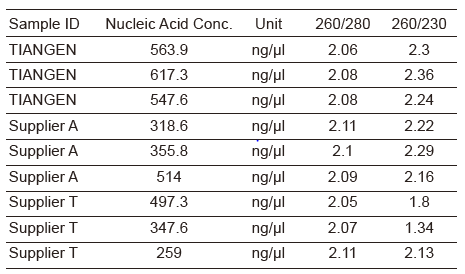
To assess the performance of the NEBNext Ultra II FS DNA library kit, the ZymoBIOMICS Microbial Community DNA Standard (Cat. No. D6306) was utilized.
This standard contains a known DNA composition of 10 microbial strains, enabling the evaluation of entire metagenomic workflows. It highlights potential biases and errors that may arise during processing, making it an ideal tool for testing library preparation kits.
It allows for a direct performance comparison with previously tested library kits at the CGR. Testing was conducted in triplicate using total input amounts of 50 ng, 1 ng, and 0.1 ng, as illustrated in Figure 2.
Figure 2. Schematic representation detailing how NEBNext libraries were generated using the ZymoBIOMICS Microbial Community DNA Standard as input DNA. Manual libraries were made using full volumes according to protocol, automated libraries were made using 1 in 10 volume on the SPT Labtech mosquito HV platform. Image Credit: SPT Labtech
Figure 3. Fragment Analyzer traces comparing the size distribution of manual and 1/10 automated libraries for A) 50 ng, B) 1 ng, and C) 0.1 ng of input DNA. Image Credit: SPT Labtech
Figure 4. Bar chart showing average yields for the manual and 1/10 automated libraries with differing input amounts and PCR cycle numbers. Error bars indicate standard deviation. Image Credit: SPT Labtech
Figure 5. Sequence data were normalized to 13M reads per library, A) indicates average levels of duplicate and mapping percentages, B) shows average fold coverage. Error bars indicate standard deviation. Image Credit: SPT Labtech
Although the ZymoBIOMICS Microbial Community DNA Standard yielded promising results, it is not always indicative of performance with real-world samples.
Previously generated data from clinical bacterial isolates utilized manually prepared TruSeq Nano libraries with 100 ng of input DNA. The same samples were subjected to the automated 1 in 10 volume NEBNext Ultra II FS DNA method with 10 ng of input DNA, and the results were compared.
Figure 6. Fragment Analyzer traces comparing the size distribution of manual TruSeq Nano and 1/10 volume automated NEBNext Ultra II FS DNA libraries. Image Credit: SPT Labtech
Figure 7. Bar chart detailing average yields for the manual TruSeq Nano and 1/10 automated NEB libraries generated from clinical bacterial isolates. 100 ng and 10 ng of input DNA were used for the TruSeq Nano and NEB 1/10 libraries, and 8 and 10 cycles of PCR, respectively. Error bars are standard deviation. Image Credit: SPT Labtech
Figure 8. Sequence data were normalized to 1.3M reads per library. Bar chart shows averages of duplicate and mapping percentages, and fold coverage of the genome. Error bars are standard deviation. Image Credit: SPT Labtech
Previously, workflows in the CGR used Illumina TruSeq Nano or TruSeq PCR Free libraries. Data was generated using these methods, both manually and with Beckman full volume automation on the FXP system.
Input included 100 ng and 1 µg of the ZymoBIOMICS Microbial Community DNA Standard for TruSeq Nano and TruSeq PCR Free libraries, respectively. Data demonstrated comparability across all three methods when 50 ng of DNA was employed as input for the 1 in 10 volume NEBNext Ultra II FS DNA libraries.
Figure 9. Sequence data were normalized to 13M reads per library. Bar chart shows averages of A) percentage duplicates, B) percentage mapping, and C) fold coverage (X) of the mock community. Error bars are standard deviation. Image Credit: SPT Labtech
This article demonstrates that NEBNext Ultra II FS DNA libraries prepared at 1 in 10 performed as well as the full volume manually prepared libraries while providing significant cost savings through the miniaturization of reaction volumes.
Even with input DNA as low as 1 ng, comparable percentages of duplicates, mapping levels, fold coverage, and GC skew (not shown here) were observed. The entire workflow can be completed in a single day, facilitating larger projects with more samples and reduced technical bias.
SPT Labtech designs and manufactures robust, reliable and easy-to-use solutions for life science. We enable life scientists through collaboration, deep application knowledge, and leading engineering to accelerate research and make a difference together. We offer a portfolio of products within sample management, liquid handling, and multiplexed detection that minimize assay volumes, reduce material handling costs and put the discovery tools back in the hands of the scientist.
Many of our innovations have been born out of the desire to create solutions to existing customer problems; and it’s this ethos that drives SPT Labtech’s R&D efforts. Our strengths come from the trust our customers have with us to develop truly unique, automated technologies to meet their needs. We combine cutting edge science with first-rate engineering to put customers at the heart of everything we do.
The substantial breadth of expertise within our company enables us to be involved in the full life cycle of our products from the initial design concept, mechanical and software engineering and prototyping, to final manufacture and sale. These qualities allow us to offer the best possible technical and mechanical support to all the equipment that we supply, hence maintaining excellent client relationships.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last updated: Oct 23, 2023 at 9:50 AM
Please use one of the following formats to cite this article in your essay, paper or report:
SPT Labtech. (2023, October 23). Investigating the miniaturization of DNA library prep for Illumina sequencing. News-Medical. Retrieved on November 08, 2023 from https://www.news-medical.net/whitepaper/20231023/Investigating-the-miniaturization-of-DNA-library-prep-for-Illumina-sequencing.aspx.
SPT Labtech. "Investigating the miniaturization of DNA library prep for Illumina sequencing". News-Medical. 08 November 2023. <https://www.news-medical.net/whitepaper/20231023/Investigating-the-miniaturization-of-DNA-library-prep-for-Illumina-sequencing.aspx>.
SPT Labtech. "Investigating the miniaturization of DNA library prep for Illumina sequencing". News-Medical. https://www.news-medical.net/whitepaper/20231023/Investigating-the-miniaturization-of-DNA-library-prep-for-Illumina-sequencing.aspx. (accessed November 08, 2023).
SPT Labtech. 2023. Investigating the miniaturization of DNA library prep for Illumina sequencing. News-Medical, viewed 08 November 2023, https://www.news-medical.net/whitepaper/20231023/Investigating-the-miniaturization-of-DNA-library-prep-for-Illumina-sequencing.aspx.
Cells dispensed by automated dispensers shown to exhibit normal apoptotic responses
Exploring assay miniaturization with positive-displacement liquid handlers
Exploring the miniaturized magnetic bead cleanup process
Harnessing the power of pipettors for accessible automation and research
How can we achieve the most effective laboratory sample management?
Improving the quality of drug discovery with low-volume workflows
Next-generation sequencing (NGS) to empower scientists in the genomic revolution
Overcoming the challenges of liquid handling for laboratory developed tests
Using an automated liquid dispenser for robust and gentle cell plating
Using automated liquid handling for efficient lab developed testing
Utilizing liquid handlers in Nextera XT library preparation
Why do we need automation-friendly library prep when it comes to handling precious samples?
News-Medical.Net provides this medical information service in accordance with these terms and conditions. Please note that medical information found on this website is designed to support, not to replace the relationship between patient and physician/doctor and the medical advice they may provide.
News-Medical.net - An AZoNetwork Site
Coptic Egyptian Dna Owned and operated by AZoNetwork, © 2000-2023